|
chr21_+_45699830
|
9.651
|
NM_030582
NM_130444
|
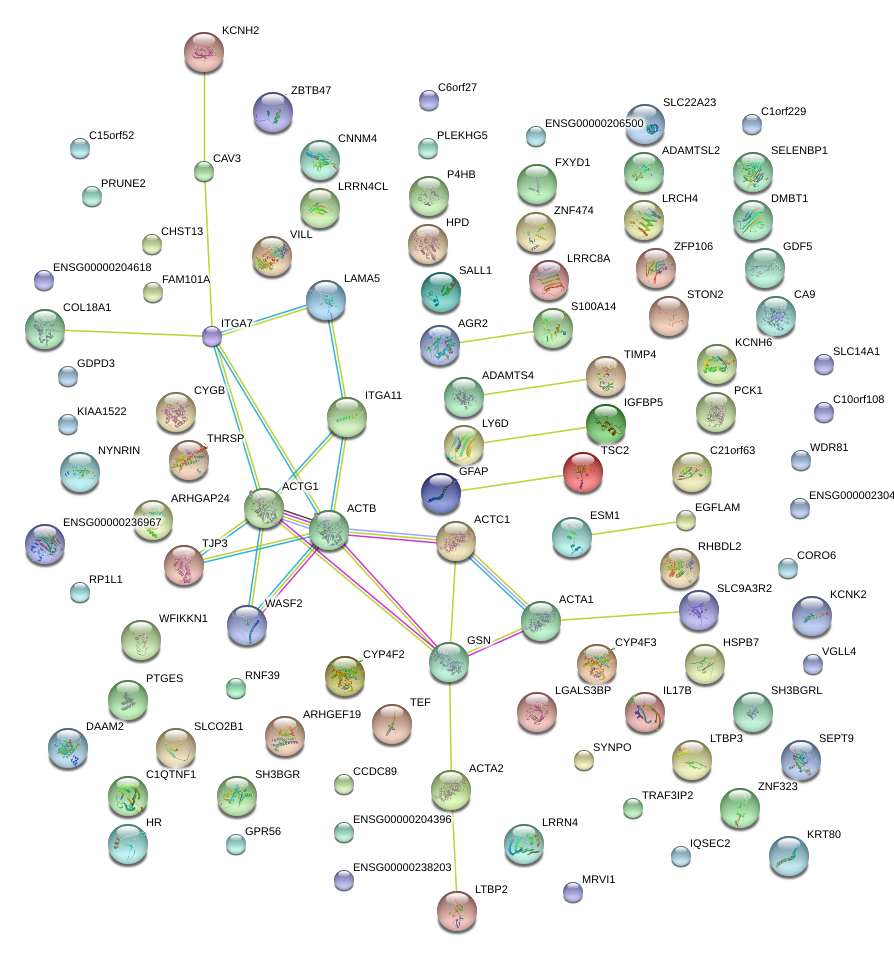
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr22_+_40093245
|
8.556
|
NM_001145398
|
TEF
|
thyrotrophic embryonic factor
|
|
chr17_-_77394872
|
8.092
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr17_+_1580504
|
6.515
|
|
WDR81
|
WD repeat domain 81
|
|
chr17_+_74542066
|
6.470
|
NM_198593
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr16_-_30032330
|
6.257
|
NM_024307
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3
|
|
chr2_-_217268492
|
6.198
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr12_-_54392328
|
6.190
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr19_+_15612706
|
4.100
|
NM_000896
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3
|
|
chr1_-_16217025
|
4.007
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr15_-_38420414
|
3.891
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr11_-_62213577
|
3.803
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr8_-_22044463
|
3.754
|
NM_005144
NM_018411
|
HR
|
hairless homolog (mouse)
|
|
chrX_-_53327473
|
3.695
|
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr2_+_96818048
|
3.564
|
|
CNNM4
|
cyclin M4
|
|
chr7_-_16811131
|
3.531
|
NM_006408
|
AGR2
|
anterior gradient homolog 2 (Xenopus laevis)
|
|
chr12_-_50871953
|
3.457
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr9_+_35663852
|
3.448
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr17_-_74487527
|
3.407
|
NM_005567
|
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein
|
|
chr9_+_139241838
|
3.386
|
NM_001190228
|
LOC643596
|
hypothetical LOC643596
|
|
chr20_-_60360789
|
3.379
|
|
LAMA5
|
laminin, alpha 5
|
|
chr5_+_38481392
|
3.328
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr5_+_150000394
|
3.318
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr11_-_62213946
|
3.253
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr3_+_8750485
|
3.092
|
NM_033337
NM_001234
|
CAV3
|
caveolin 3
|
|
chr17_+_75507750
|
3.085
|
|
|
|
|
chr11_-_85074848
|
3.083
|
NM_152723
|
CCDC89
|
coiled-coil domain containing 89
|
|
chr15_-_38417564
|
3.061
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr9_+_130719034
|
3.019
|
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr16_+_620932
|
3.002
|
NM_053284
|
WFIKKN1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1
|
|
chr17_-_24972471
|
2.941
|
NM_032854
|
CORO6
|
coronin 6
|
|
chr3_+_127743868
|
2.787
|
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13
|
|
chr1_+_213245507
|
2.783
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr11_-_65082705
|
2.718
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr8_-_10550026
|
2.687
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr8_-_143864933
|
2.680
|
NM_003695
|
LY6D
|
lymphocyte antigen 6 complex, locus D
|
|
chr6_-_30151526
|
2.673
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr1_-_39180036
|
2.610
|
NM_017821
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila)
|
|
chr20_+_55569542
|
2.577
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr3_+_38007264
|
2.546
|
|
VILL
|
villin-like
|
|
chr3_-_11585264
|
2.545
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr16_+_2071631
|
2.538
|
|
TSC2
|
tuberous sclerosis 2
|
|
chr7_-_150283794
|
2.476
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr6_-_112001590
|
2.473
|
NM_001164282
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr5_-_148738980
|
2.471
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr15_-_32875040
|
2.461
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr19_+_40322231
|
2.442
|
NM_005031
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr11_-_10672079
|
2.421
|
NM_001100163
NM_130385
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr17_+_72795567
|
2.384
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr1_+_33003814
|
2.363
|
NM_001198972
NM_001198973
|
KIAA1522
|
KIAA1522
|
|
chr20_-_5982617
|
2.362
|
NM_152611
|
LRRN4
|
leucine rich repeat neuronal 4
|
|
chr12_+_123339662
|
2.359
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr1_-_16411690
|
2.348
|
NM_153213
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19
|
|
chr12_-_120781002
|
2.332
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr14_-_80934523
|
2.295
|
NM_033104
|
STON2
|
stonin 2
|
|
chr6_+_39868770
|
2.285
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr1_-_6502655
|
2.188
|
NM_198681
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr11_+_74539679
|
2.140
|
NM_001145212
NM_007256
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr1_+_33003847
|
2.134
|
|
KIAA1522
|
KIAA1522
|
|
chr11_+_77452550
|
2.077
|
NM_003251
|
THRSP
|
thyroid hormone responsive
|
|
chr9_-_131555111
|
2.053
|
NM_004878
|
PTGES
|
prostaglandin E synthase
|
|
chr21_+_39745649
|
2.053
|
NM_007341
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein
|
|
chr6_-_31853027
|
2.051
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr18_+_41558089
|
1.987
|
NM_001128588
NM_001146036
NM_015865
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr3_-_12175339
|
1.986
|
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr1_-_245342150
|
1.986
|
NM_207401
|
C1orf229
|
chromosome 1 open reading frame 229
|
|
chr1_-_159435440
|
1.983
|
NM_005099
|
ADAMTS4
|
ADAM metallopeptidase with thrombospondin type 1 motif, 4
|
|
chr11_-_10671736
|
1.963
|
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr16_+_56219802
|
1.961
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr14_+_23937766
|
1.937
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr17_-_40348378
|
1.914
|
NM_001131019
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr11_-_65082208
|
1.909
|
NM_001130144
NM_001164266
NM_021070
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr17_-_72045218
|
1.898
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr11_-_10671873
|
1.896
|
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr20_-_33489383
|
1.893
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr21_+_32706615
|
1.843
|
NM_058187
|
C21orf63
|
chromosome 21 open reading frame 63
|
|
chr9_+_135389698
|
1.821
|
NM_014694
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr1_-_151855413
|
1.808
|
NM_020672
|
S100A14
|
S100 calcium binding protein A14
|
|
chr9_-_78710698
|
1.807
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr15_-_66511488
|
1.773
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr10_+_685887
|
1.763
|
|
C10orf108
|
chromosome 10 open reading frame 108
|
|
chr10_+_124310170
|
1.728
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr3_+_42675841
|
1.719
|
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr1_-_149611768
|
1.706
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr19_+_3659358
|
1.689
|
|
TJP3
|
tight junction protein 3 (zona occludens 3)
|
|
chr6_-_3390197
|
1.678
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr16_-_49742651
|
1.657
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr16_+_2025922
|
1.646
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr18_+_41558141
|
1.640
|
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr8_-_13178327
|
1.614
|
|
|
|
|
chr4_+_86968069
|
1.576
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr9_+_123083540
|
1.574
|
|
GSN
|
gelsolin
|
|
chr5_-_54317102
|
1.571
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr6_-_28429950
|
1.569
|
NM_145909
|
ZNF323
|
zinc finger protein 323
|
|
chr1_-_27604656
|
1.525
|
|
WASF2
|
WAS protein family, member 2
|
|
chr5_+_121493113
|
1.505
|
NM_207317
|
ZNF474
|
zinc finger protein 474
|
|
chr17_-_39265965
|
1.503
|
NM_001932
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3)
|
|
chr1_-_151630168
|
1.495
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr22_+_31526801
|
1.495
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr1_+_159435728
|
1.473
|
NM_004550
|
NDUFS2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase)
|
|
chr20_-_43601542
|
1.466
|
NM_080827
|
WFDC6
|
WAP four-disulfide core domain 6
|
|
chr1_-_218168548
|
1.457
|
NM_018713
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr17_-_36396778
|
1.450
|
NM_182497
|
KRT40
|
keratin 40
|
|
chr12_+_16397617
|
1.407
|
NM_145764
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr1_+_1207351
|
1.395
|
NM_001130413
NM_002978
|
SCNN1D
|
sodium channel, nonvoltage-gated 1, delta
|
|
chr20_+_58004811
|
1.394
|
NM_021810
|
CDH26
|
cadherin 26
|
|
chrX_+_99785818
|
1.389
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr8_-_13018123
|
1.381
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr19_-_40696393
|
1.366
|
NM_001126056
NM_001126057
NM_001126058
NM_001190347
NM_001190348
NM_001190349
NM_033317
|
DMKN
|
dermokine
|
|
chr17_+_71065478
|
1.355
|
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr1_+_61319976
|
1.350
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr17_-_44226595
|
1.345
|
NM_173623
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6
|
|
chr19_+_51493486
|
1.314
|
NM_022462
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr19_+_50004134
|
1.308
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr14_+_85066207
|
1.295
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr9_-_35860316
|
1.292
|
NM_001004487
|
OR13J1
|
olfactory receptor, family 13, subfamily J, member 1
|
|
chr19_+_51498695
|
1.276
|
NM_152794
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr16_+_55963899
|
1.264
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr2_-_11727705
|
1.260
|
NM_012344
|
NTSR2
|
neurotensin receptor 2
|
|
chr8_+_22900863
|
1.245
|
NM_001160036
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr9_+_138991835
|
1.236
|
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr19_-_4406285
|
1.215
|
NM_001171091
|
UBXN6
|
UBX domain protein 6
|
|
chr1_+_116181927
|
1.209
|
|
NHLH2
|
nescient helix loop helix 2
|
|
chr1_+_183970093
|
1.209
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr1_+_1836125
|
1.193
|
NM_138705
|
CALML6
|
calmodulin-like 6
|
|
chr9_-_23811842
|
1.179
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr9_-_112010233
|
1.176
|
NM_001012993
|
C9orf152
|
chromosome 9 open reading frame 152
|
|
chr3_-_12175646
|
1.171
|
NM_003256
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr22_+_19239221
|
1.170
|
|
MED15
|
mediator complex subunit 15
|
|
chr9_-_23811808
|
1.158
|
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr19_-_53950458
|
1.144
|
NM_000148
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group)
|
|
chr20_+_34206070
|
1.144
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr1_-_151852128
|
1.139
|
NM_080388
|
S100A16
|
S100 calcium binding protein A16
|
|
chr9_+_138991775
|
1.126
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr16_+_750765
|
1.122
|
NM_005823
NM_001177355
|
MSLN
|
mesothelin
|
|
chr14_+_94117545
|
1.121
|
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr1_-_32036802
|
1.118
|
|
SPOCD1
|
SPOC domain containing 1
|
|
chr8_-_22841173
|
1.117
|
NM_144962
|
PEBP4
|
phosphatidylethanolamine-binding protein 4
|
|
chr1_-_227636462
|
1.110
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chr14_+_22581215
|
1.108
|
NM_001099780
|
PSMB11
|
proteasome (prosome, macropain) subunit, beta type, 11
|
|
chrX_+_153423652
|
1.105
|
NM_001099856
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr1_-_32036858
|
1.098
|
|
SPOCD1
|
SPOC domain containing 1
|
|
chr4_-_66218777
|
1.096
|
|
EPHA5
|
EPH receptor A5
|
|
chr1_+_158636987
|
1.095
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr11_-_102219450
|
1.090
|
NM_002422
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase)
|
|
chr16_-_28457825
|
1.088
|
NM_001042483
NM_012385
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr7_+_1094248
|
1.086
|
NM_001098201
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr2_+_223244606
|
1.084
|
NM_058165
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1
|
|
chr16_+_23673448
|
1.069
|
NM_022097
|
CHP2
|
calcineurin B homologous protein 2
|
|
chr9_+_27099138
|
1.066
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr5_-_172130766
|
1.064
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr19_-_360138
|
1.055
|
NM_001136263
|
C2CD4C
|
C2 calcium-dependent domain containing 4C
|
|
chr1_+_175407153
|
1.054
|
NM_021165
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr5_+_176449156
|
1.054
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr14_-_22515725
|
1.044
|
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr7_+_150376537
|
1.043
|
NM_004769
NM_020321
NM_020322
|
ACCN3
|
amiloride-sensitive cation channel 3
|
|
chr12_-_8706637
|
1.043
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr14_+_85066265
|
1.042
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chrX_-_132947141
|
1.039
|
|
GPC3
|
glypican 3
|
|
chr10_+_88708267
|
1.029
|
NM_003087
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1)
|
|
chr2_+_85515428
|
1.021
|
NM_198482
|
SH2D6
|
SH2 domain containing 6
|
|
chr1_+_151867483
|
1.015
|
NM_006271
|
S100A1
|
S100 calcium binding protein A1
|
|
chr16_+_66476281
|
1.014
|
NM_198443
|
NRN1L
|
neuritin 1-like
|
|
chrX_+_2756810
|
1.004
|
NM_001079855
NM_001184702
NM_001184703
NM_001184704
NM_003918
|
GYG2
|
glycogenin 2
|
|
chr5_+_1254709
|
0.989
|
NM_001003841
|
SLC6A19
|
solute carrier family 6 (neutral amino acid transporter), member 19
|
|
chrX_+_122922058
|
0.982
|
NM_001042749
|
STAG2
|
stromal antigen 2
|
|
chr1_-_218168404
|
0.978
|
|
|
|
|
chr2_+_26924466
|
0.974
|
NM_020134
|
DPYSL5
|
dihydropyrimidinase-like 5
|
|
chr8_-_62764839
|
0.974
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr6_-_119512035
|
0.972
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr1_-_159325709
|
0.966
|
|
PVRL4
|
poliovirus receptor-related 4
|
|
chrX_+_46822751
|
0.958
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr6_-_39305203
|
0.951
|
NM_003740
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr12_+_99391681
|
0.948
|
NM_005123
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr10_+_135010833
|
0.947
|
NM_001145201
NM_145202
|
PRAP1
|
proline-rich acidic protein 1
|
|
chr10_-_128067052
|
0.947
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr22_-_29290875
|
0.946
|
NM_004861
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1
|
|
chr22_+_43451351
|
0.943
|
NM_001017530
NM_001198721
NM_015366
|
PRR5
|
proline rich 5 (renal)
|
|
chr8_+_24828359
|
0.942
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr12_+_118100977
|
0.940
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr11_+_67532592
|
0.937
|
NM_001161473
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr17_-_64462976
|
0.936
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chrX_-_132947290
|
0.936
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr10_+_80743769
|
0.927
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr1_-_36338436
|
0.923
|
NM_005202
|
COL8A2
|
collagen, type VIII, alpha 2
|
|
chr1_-_203657803
|
0.921
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr4_+_183482130
|
0.921
|
NM_001080477
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr9_+_6205804
|
0.919
|
|
IL33
|
interleukin 33
|
|
chr11_-_35397675
|
0.917
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr2_+_215884923
|
0.914
|
NM_004044
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chrX_+_106057984
|
0.905
|
|
CLDN2
|
claudin 2
|
|
chr10_-_128066935
|
0.904
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr1_-_27089354
|
0.903
|
|
GPN2
|
GPN-loop GTPase 2
|
|
chr5_-_59819642
|
0.901
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_52618815
|
0.901
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr3_-_12561962
|
0.898
|
NM_001195279
|
LOC100129480
|
hypothetical protein LOC100129480
|
|
chr22_-_37970902
|
0.896
|
NM_002608
|
PDGFB
|
platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog)
|
|
chr1_+_238321794
|
0.895
|
NM_020066
|
FMN2
|
formin 2
|
|
chr1_-_240679380
|
0.895
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr12_+_56406296
|
0.895
|
|
LOC100130776
|
hypothetical LOC100130776
|